Scientific and software engineering examples of applied category theory
Kris Brown - Topos Seminar

(press s for speaker notes)
6/5/23
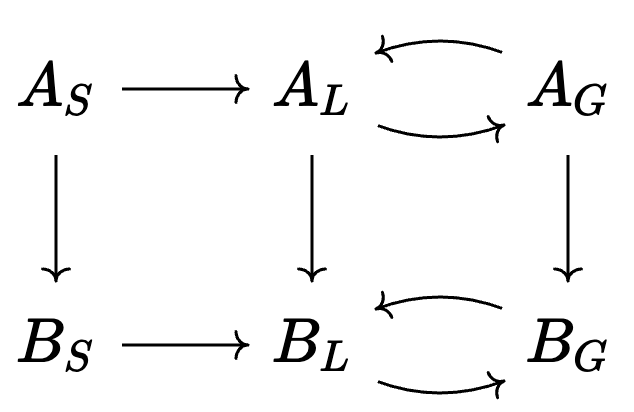
Abstraction
$$
$$
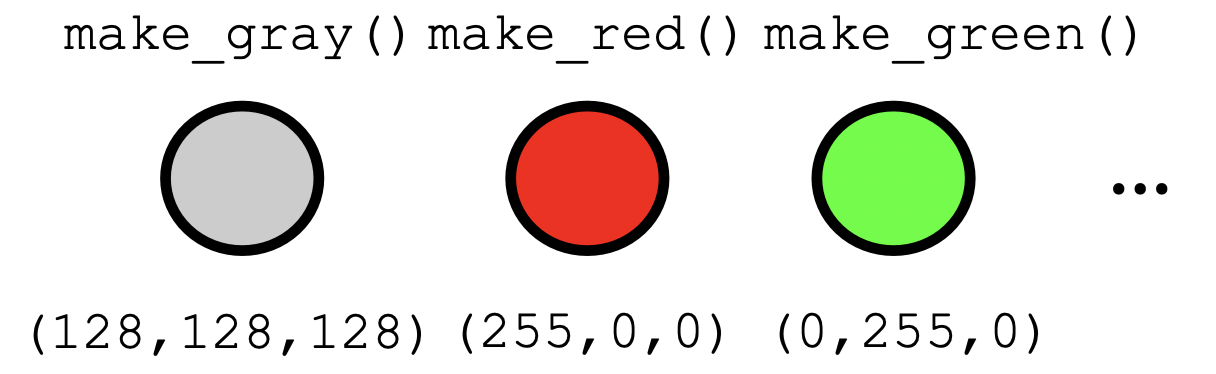
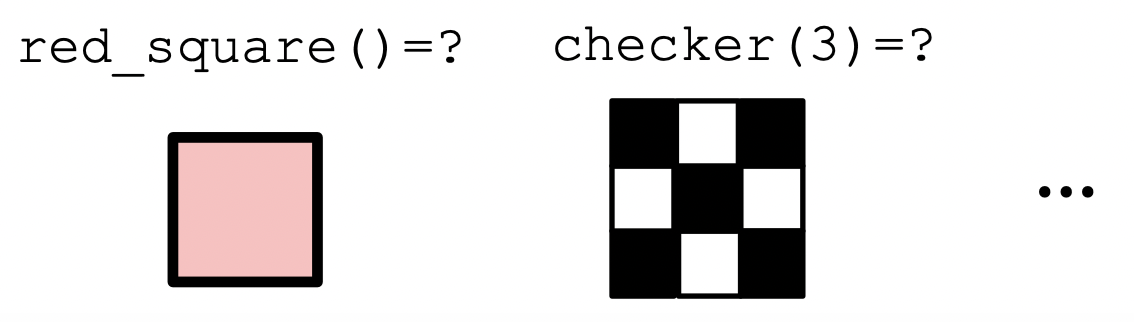
The status quo
Abstraction is something all programmers are familiar with.
- There are pros and cons to it. Cons follow when abstractions are ad hoc.
- E.g. they don’t fit well together, difficult to modify, unintelligible to peers
Category theory offers abstractions that fit well together.
- Scientific workflow can be improved to be conceptual, not manual, programming.
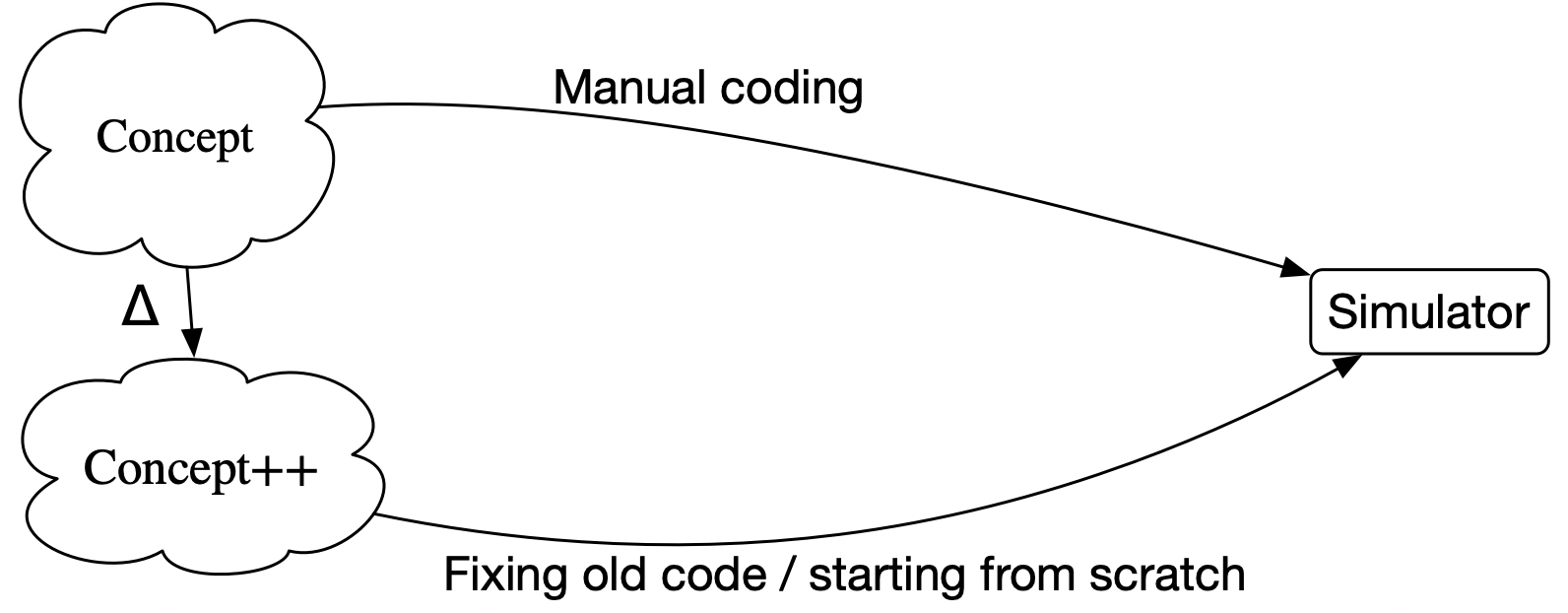
A better quo
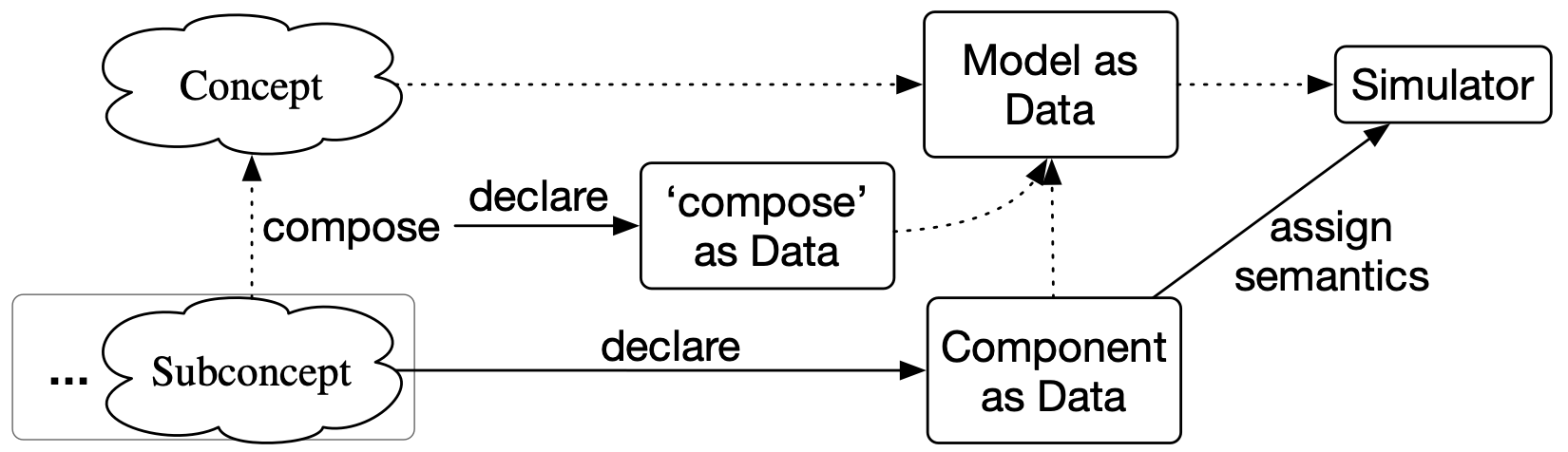
⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅
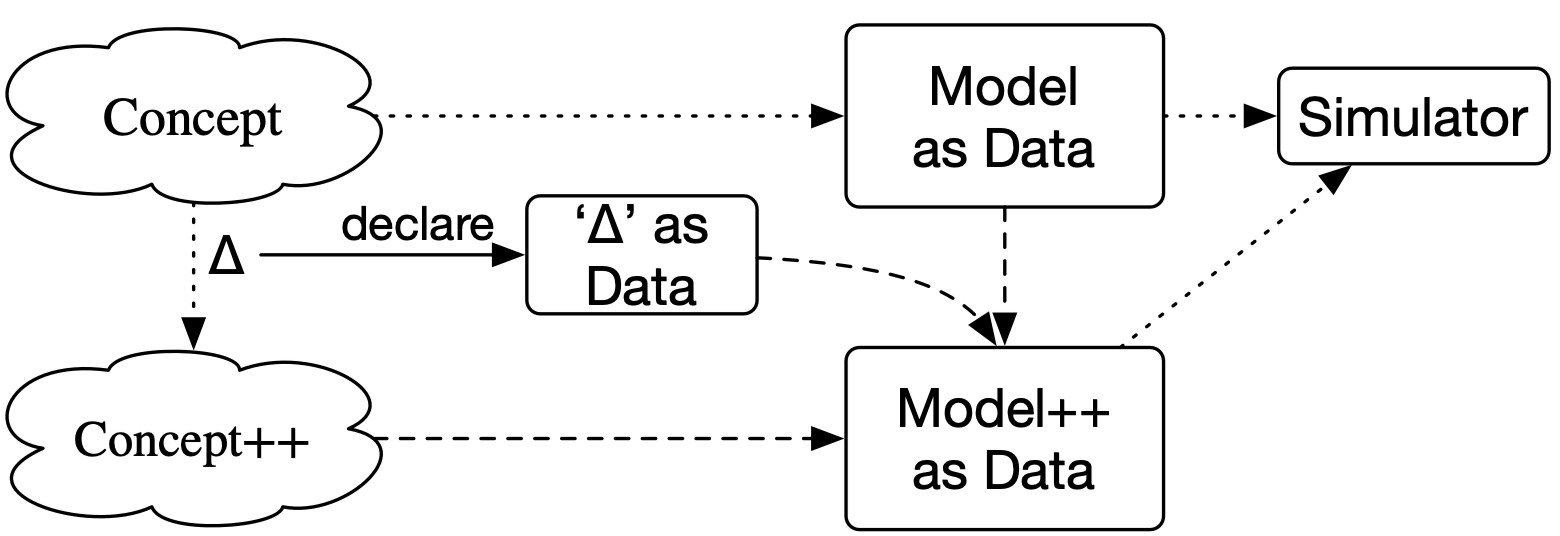
Why Category Theory?
Focuses on relationships between things without talking about the things themselves.
Invented in the 1940’s to connect different branches of math.
A category consists of objects and morphisms (arrows).
- We don’t need to know anything about the objects.
- Compose \(A \rightarrow B\) and \(B \rightarrow C\) to get \(A \rightarrow C\).
- Like a graph, but we care about paths, not edges.
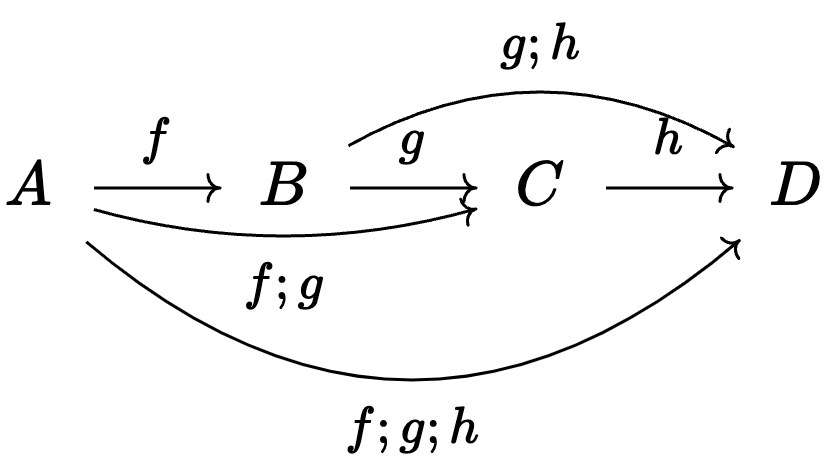
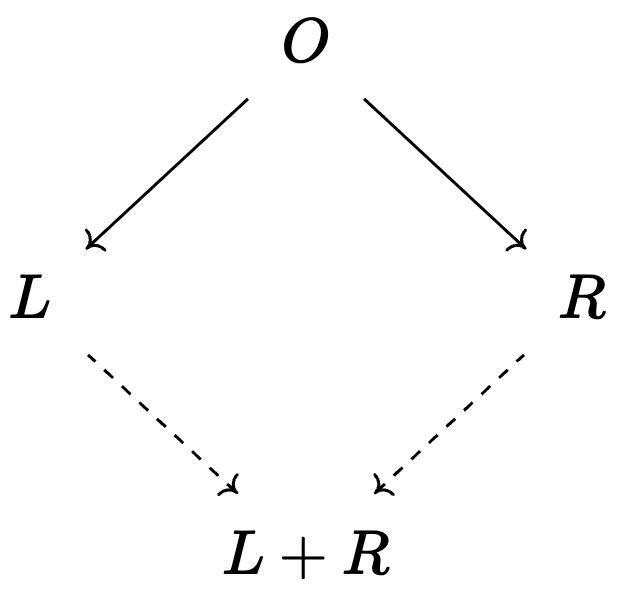
CT studies certain shapes of combinations of arrows.
- These can be local shapes, e.g. a span: \(\huge \cdot \leftarrow \cdot \rightarrow \cdot\)
- These can be global, e.g. an initial object: \(\huge \boxed{\cdot \rightarrow \cdot\rightarrow \cdot \rightarrow \dots}\)
Applied Category Theory?
Compare to interfaces in computer science:
- declare that some collection of things are related in a particular way without saying what they are.
In some sense a category is just a particular interface.
- Category of sets and functions
- Category of sets and subsets
- Category of \(\mathbb{Z}\) and \(\leq\)
- Category of categories and functors
- Category of chemical reaction networks
- Category of chemical structures
- Category of datasets
- Category of datatypes and programs
CT is also the study of interfaces in general. It knows which are good ones.
Outline
- 🗄️ Data
- CT: C-Sets, Homomorphisms, Data migration
- Examples: Computational chemistry simulation generated data
- 💭 Models
- CT: Colimits, Limits
- Examples: COVID models, Chemical Reaction Networks
- 📈 Simulation
- CT: Functors, DWDs, rewriting
- Examples: Multiphysics PDEs, Agent-based models
🗄️. DFT simulations
A simulation has the following data
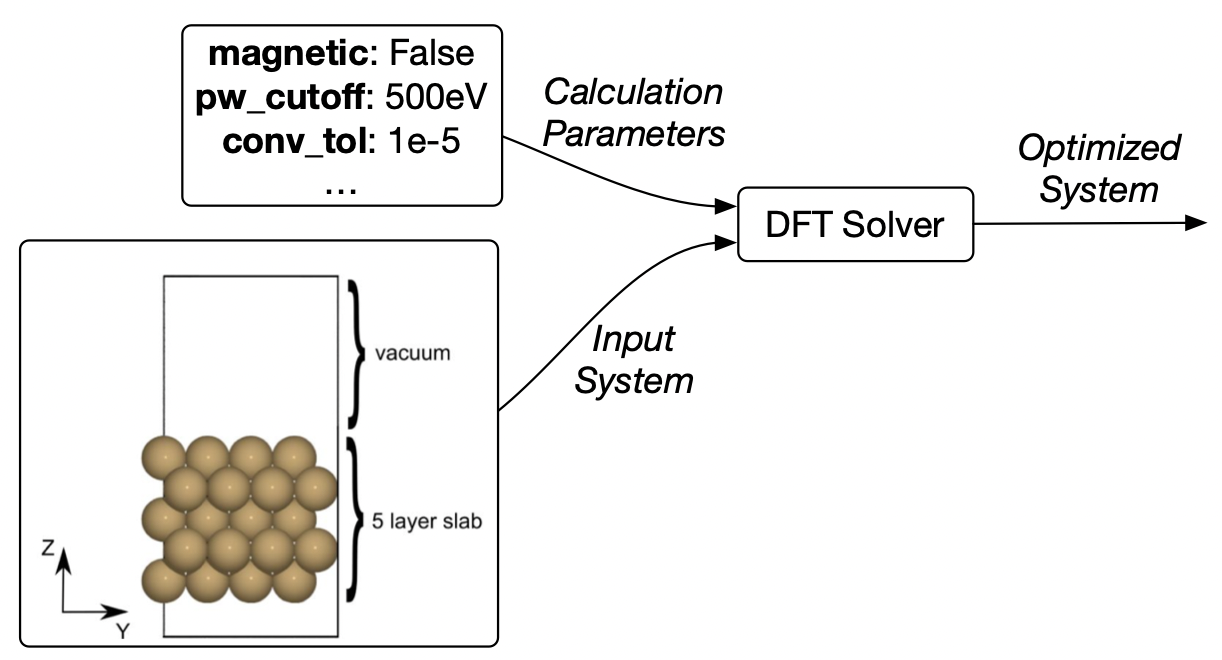
🗄️. Storing the data of lots of simulations
simulations
|-bulk
| |-magnetic
| | |- Fe
| | | |-pw_500/
| | | |-pw_600/
| | | |-pw_700/
| |-nonmagnetic
| | |- Al
| | | |-pw_500/
| | | |-pw_700/
|-surface
| |-...
|-random
| |- Ni lattice constant/
| |- OH adsorbate interaction/Problem
- Structure is not made explicit (unconstrained)
- It’s the wrong structure
- arbitrary order on parameters
- duplication of work
Instead, assemble the simulations into a big list and deduce experiments:
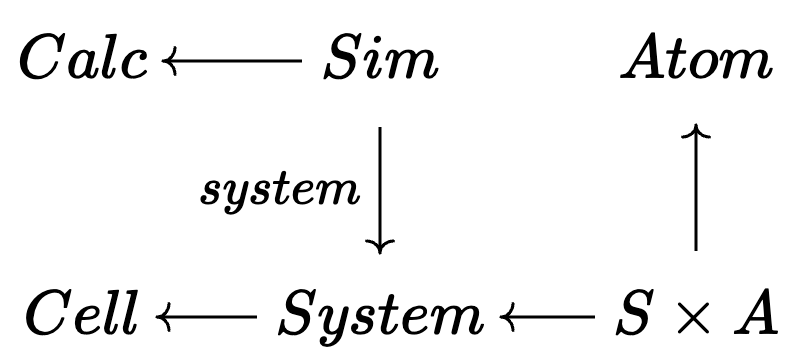
class Simulation():
def __init__(self, calc: Calculator, sys: System):
def write_input(self, pth: str):
def read_input(pth: str) -> Simulation:
class Calculator():
def __init__(self, params: Dict[str, Any]):
class System():
def __init__(self, cell:Cell, atoms:List[Atom]):
class Cell():
def __init__(self, dims:np.ndarray, pbc=Tuple[Bool,Bool,Bool]):
class Atom(Species):
def __init__(self, elem: str,x:float,y:float,z:float):Problem
- Inefficient/tedious querying
- Inefficient/tedious serialization
- Fragile infrastructure
🗄️. An improvement: databases
Relational databases
- Each has a schema of entities, foreign keys, and attributes.
- These allow one to model things, relationships, and properties.
- A database instance has:
- a table for each entity
- a column for each FK+Attr
- a table for each entity
- Instances be efficiently queried using SQL language.
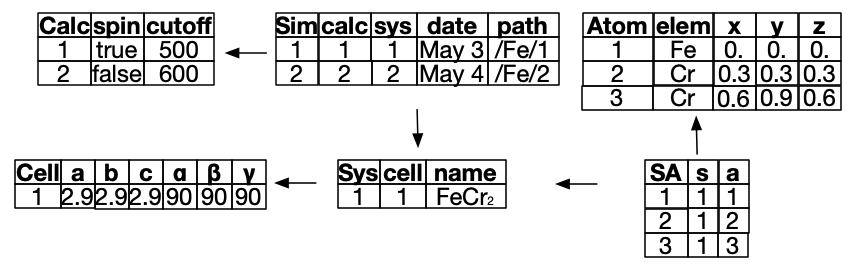
Example instance
🗄️. Another improvement: C-Sets / ACSets
Problem
SQL solves the issue of querying and storage of data. But what do we do when we need to do more?
We must convert back from the database back into the custom data type, dealing with all of the problems associated with that.
A \(\mathsf{C}\)-set has the same information as a database without attributes.
Attributed \(\mathsf{C}\)-sets (ACSets) have the same information as ordinary relational databases.
Unlike databases, we understand ACSets as living in a category which supplies many interesting and useful things to do with database that we would otherwise not think to do.
The majority of this talk will emphasize the sorts of things one can do with this perspective.
Example ACSet


🗄️. Head-to-head comparison



# Python
class Atom():
def __init__(self, elem,x,y,z):
def __eq__(self, other):
class Cell():
def __init__(self, dims, pbc):
def __eq__(self, other):
class System():
def __init__(self, cell, atoms):
def __eq__(self, other):
class Calculator():
def __init__(self, params):
def __eq__(self, other):
class Sim():
def __init__(self, calc, sys):
def __eq__(self, other):
def write_input(self, pth: str):
def read_input(pth: str) -> Sim:# AlgebraicJulia
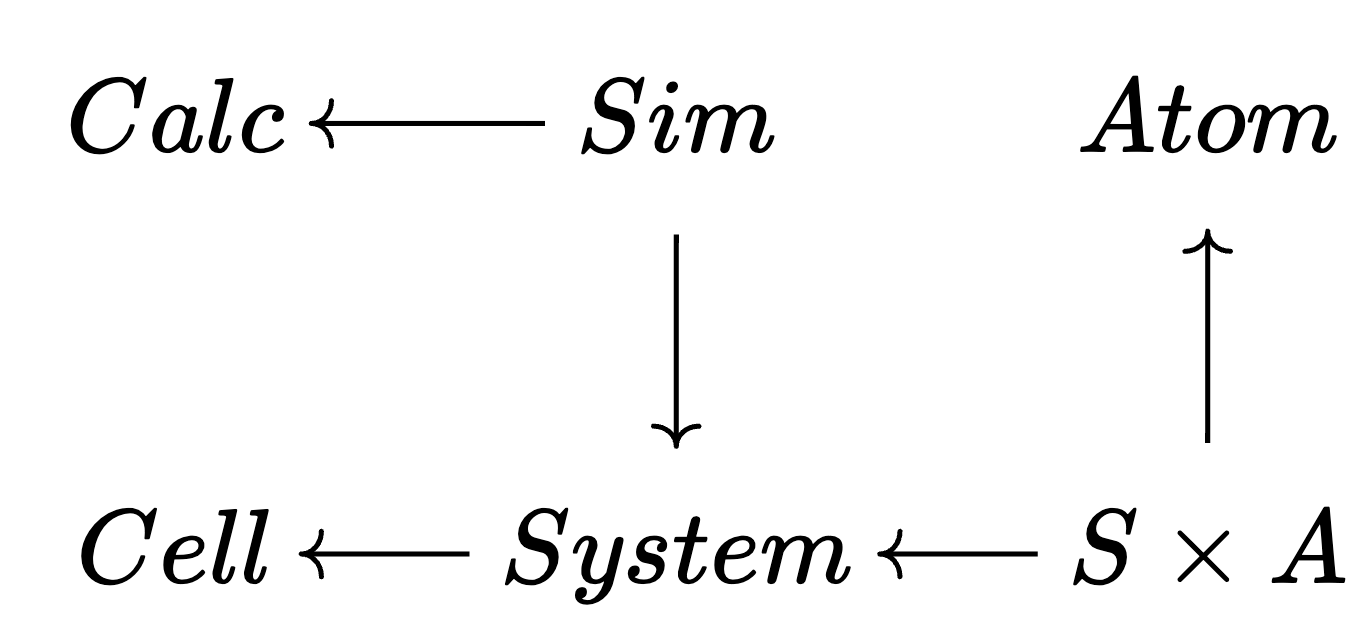
@present Simulations begin
(Atom,S_A,Cell,System,Sim,Calc) :: Ob
(String, Float, Int) :: AttrType
calc :: Hom(Sim, Calc)
system :: Hom(Sim, System)
cell :: Hom(System, Cell)
s_system :: Hom(S_A, System)
s_atom :: Hom(S_A, Atom)
pw :: Attr(Calc, Float)
xc :: Attr(Calc, String)
(x,y,z) :: Attr(Atom, Float)
elem :: Attr(Atom, Int)
endFind pairs of simulations with the same calculator and cell but different atomic configurations.
# Python
CHEM_LEVEL = 3
"""Put paths into buckets. Same bucket if
their path (*ignoring* the 3rd folder name,
i.e. chemical structure) is the same."""
def find_pairs(sim_path_root):
EQcalc = defaultdict(list)
for p in get_all_paths(sim_path_root)
# e.g. p=["bulk","mag","Fe","pw_500",...]
# pth_no_chem=["bulk","mag","pw_500",...]
pth_no_chem = p[:CHEM_LEVEL]+p[CHEM_LEVEL+1:]
bucket = (get_calc(p), pth_no_chem)
EQcalc[bucket].append(p)
end
return EqCalcCell.values()🗄️. C-Set morphisms
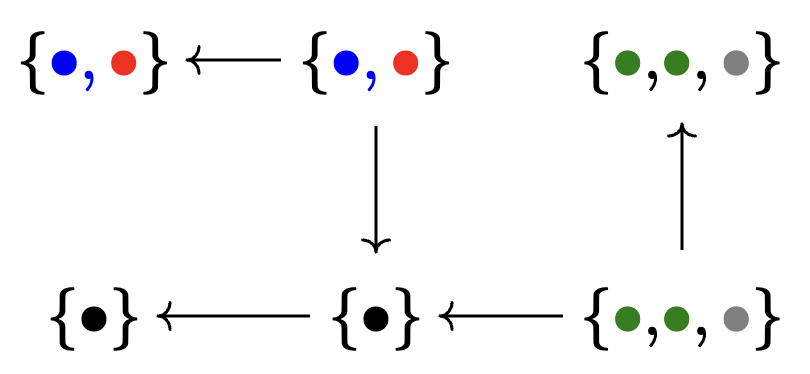
A category of ACSets and ACSet morphisms.
- A morphism \(\alpha\) has a function per entity.
- This must preserve structure, e.g. sending a \(Sim\) somewhere and looking at its \(Calc\) should be the same place where you send the \(Calc\) of that \(Sim\).
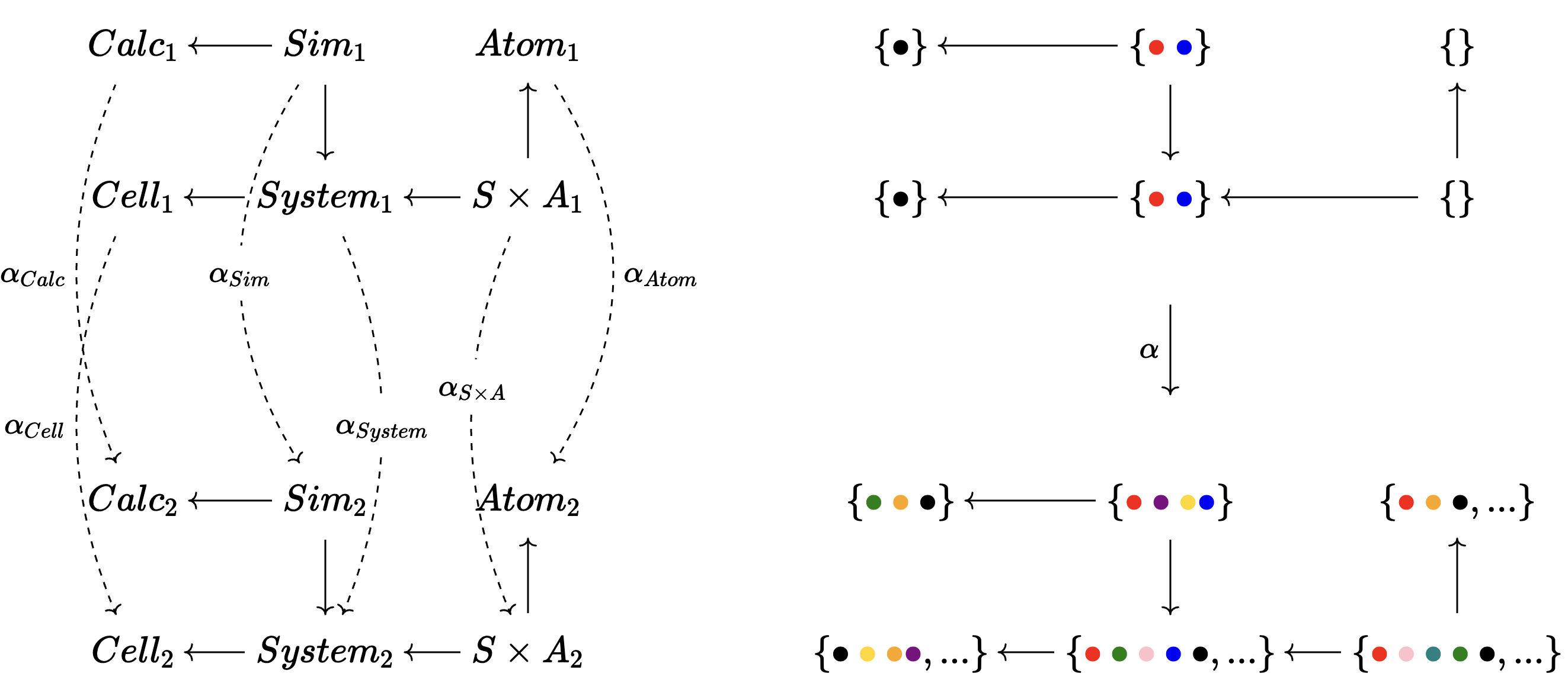
🗄️. Data migration
\[\overset{\Sigma}\Rightarrow\]
\[\longrightarrow\]
\[\underset{\Delta}\Leftarrow\]
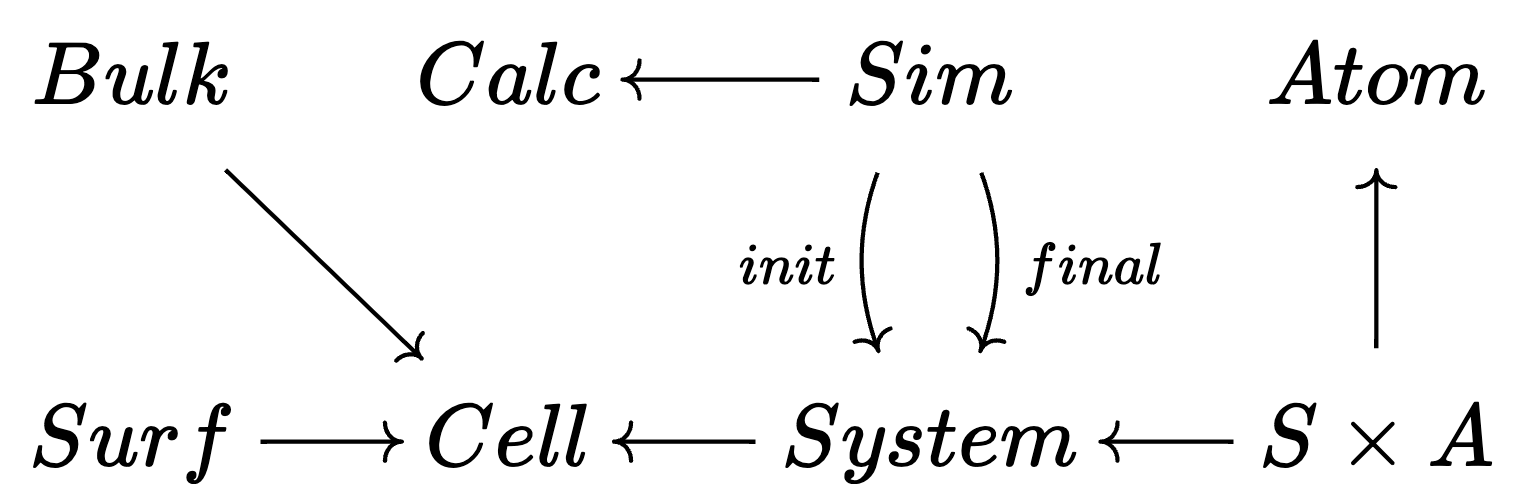
When you design a schema to contain your data, you will soon learn it needs to be updated. This is a huge problem.
Schemas are actually categories; functors between them automatically induce data migrations on arbitrary databases.
When your algorithm is expressed in terms of ACSets, then you can migrate your algorithm automatically, too!
🗄️. Data migration

\[\overset{\Sigma}\Rightarrow\]
\[\longrightarrow\]
\[\underset{\Delta}\Leftarrow\]

# Python
"""System we know nothing about (except pbc)"""
def unknown(pbc):
if pbc == [True,True,True]:
return System(Bulk(0,0,0,pbc), [])
else if pbc == [True, True, False]:
return System(Surf(0,0,0,pbc), [])
"""Migrate a Sim to a Sim2"""
def migrate_sim(sim::Sim):
init_sys = System2(sim.cell, sim.atoms)
final_sys = unknown(init_sys.cell.pbc)
return Sim2(sim.calc, init_sys, final_sys)
for old_sim in get_sims(old_db_cxn):
insert_sim(migrate_sim(old_sim), new_db_cxn)
new_query = "SELECT S1.pth, S2.pth FROM ..."# AlgebraicJulia
"""Declare relationship between schemas"""
F = FinFunctor((system=final,), Simulations, NewSimulations)
"""Constraints"""
Bb, Sb = [true,true,true], [true,true,false]
B = @acset_colim Sims begin b::Bulk; pbc(cell(b))==Bb end
S = @acset_colim Sims begin s::Surf; pbc(cell(b))==Sb end
CB = @acset Sims begin Cell=1; pbc=Bb end
CS = @acset Sims begin Cell=1; pbc=Sb end
constraints = [homomorphism(CB,B), homomorphism(CS,S)]
# Migrate old data
new_simulation_db = Σ(F,constraints)(sims)
# Migrate old query, Q
new_Q = Σ(F,constraints)(Q)🗄️. Takeaways
- Filesystem \(<\) Classes
- More flexible data model than mere tree structure
- Can write modular code for different components of data model
- Classes \(<\) Database
- Abstract away the tedium of designing efficient storage for each new structure
- Uniform language for querying the data
- Database \(<\) ACSet
- Many schema-agnostic algorithms
- Automated data migration tools
In each case, the fragility of the code you write decreases.
Outline
- 🗄️ Data
- CT: C-Sets, Homomorphisms, Data migration
- Examples: Computational chemistry simulation generated data
- 💭 Models
- CT: Colimits, Limits
- Examples: COVID models, Chemical Reaction Networks
- 📈 Simulation
- CT: Functors, DWDs, rewriting
- Examples: Multiphysics PDEs, Agent-based models
💭 Models through the eyes of the computer
Model created in AnyLogic, courtesy of Nathaniel Osgood and Xiaoyan Li
💭 Models through the eyes of the scientist
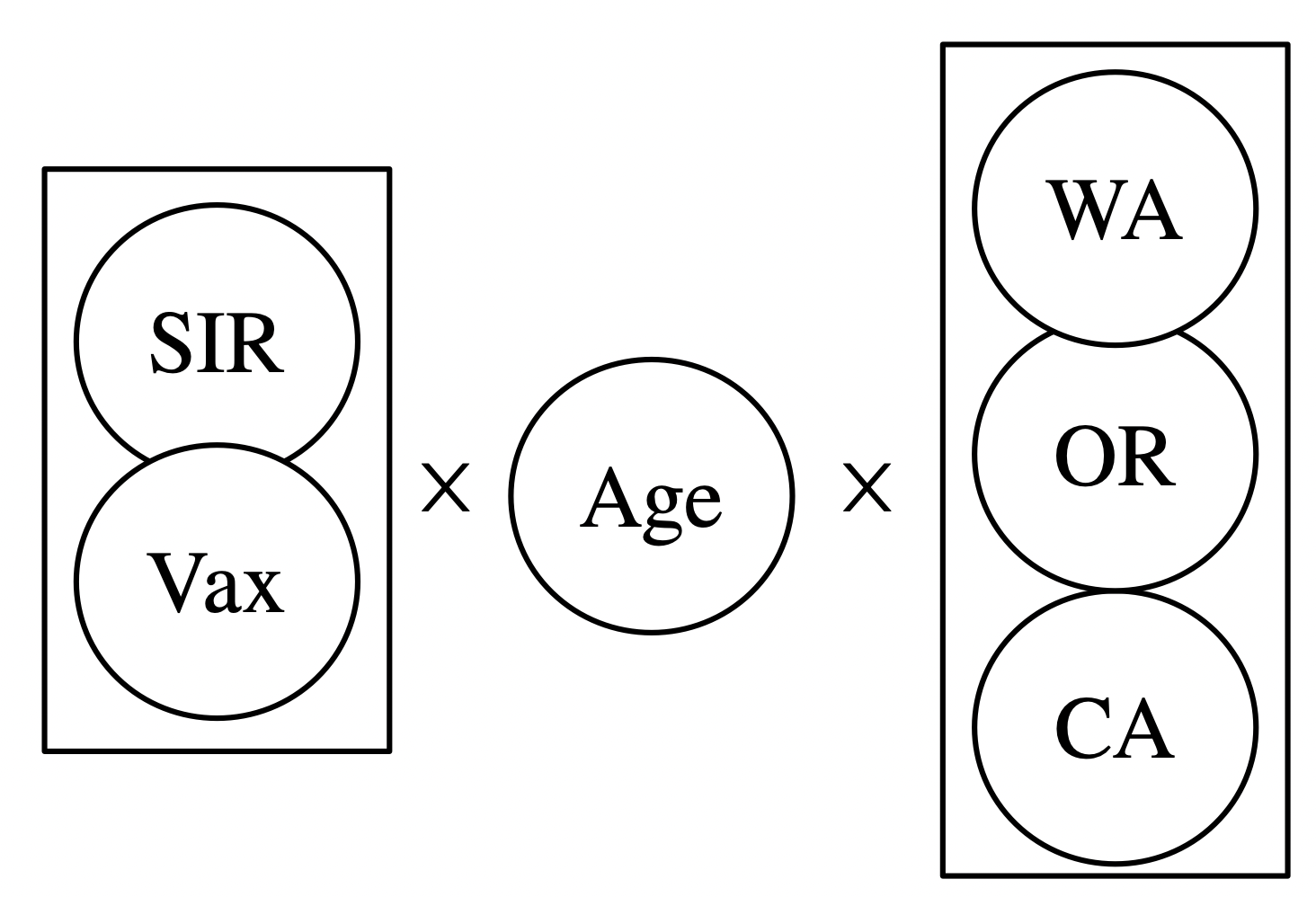
💭 C-Set for Chemical Reaction Networks
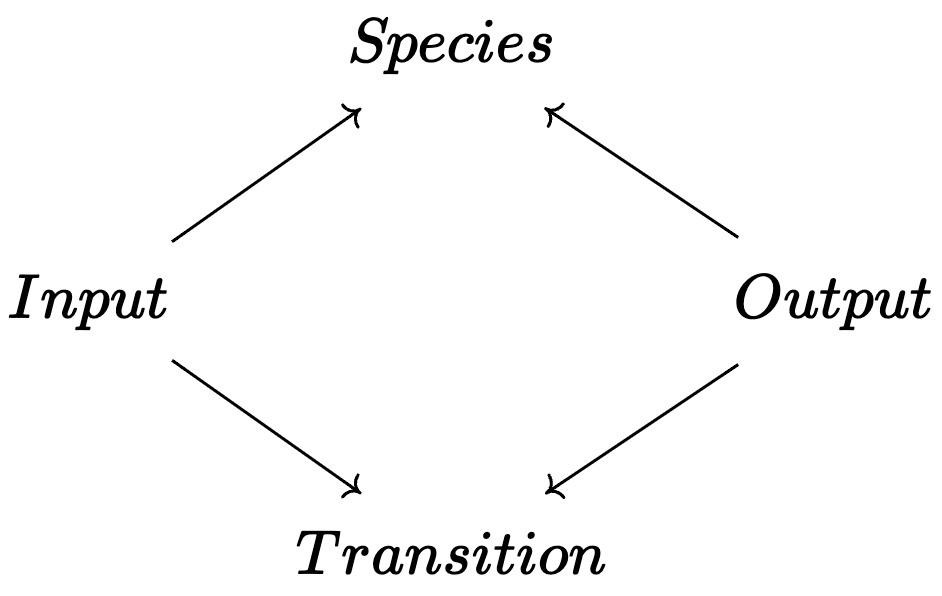
The category of Petri nets are just \(\mathsf{C}\)-Sets for a specific \(\mathsf{C}\).
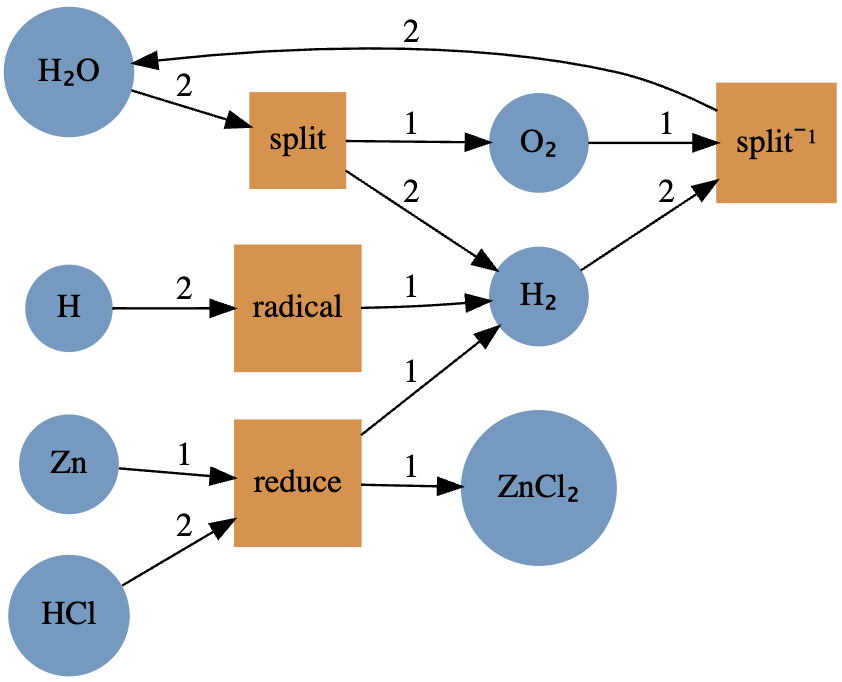
\[2\text{H}_2\text{O}\leftrightarrows 2\text{H}_2+\text{O}_2\]
\[2\text{H}\rightarrow \text{H}_2\]
\[\text{Zn}+2\text{HCl}\rightarrow \text{ZnCl}_2+\text{H}_2\]
using AlgebraicPetri
crn = LabelledPetriNet(
[:H₂O, :H₂, :O₂, :H, :Zn, :ZnCl₂, :HCl],
:split =>((:H₂O, :H₂O)=>(:H₂, :H₂, :O₂)),
:split⁻¹ =>((:H₂, :H₂, :O₂)=>(:H₂O, :H₂O)),
:radical =>((:H,:H)=>:H₂),
:reduce =>((:Zn,:HCl,:HCl)=>(:ZnCl₂,:H₂)),
)
┌───┬─────────┐
│ T │ tname │
├───┼─────────┤
│ 1 │ split │
│ 2 │ split⁻¹ │
│ 3 │ radical │
│ 4 │ reduce │
└───┴─────────┘
┌───┬───────┐
│ S │ sname │
├───┼───────┤
│ 1 │ H₂O │
│ 2 │ H₂ │
│ 3 │ O₂ │
│ 4 │ H │
│...│ ... │
└───┴───────┘
3 rows omitted┌───┬───┬───┐
│ I │it │is │
├───┼───┼───┤
│ 1 │ 1 │ 1 │
│ 2 │ 1 │ 1 │
│ 3 │ 2 │ 2 │
│ 4 │ 2 │ 2 │
│...│...│...│
└───┴───┴───┘
6 rows omitted
┌───┬───┬───┐
│ O │ot │os │
├───┼───┼───┤
│ 1 │ 1 │ 2 │
│ 2 │ 1 │ 2 │
│ 3 │ 1 │ 3 │
│ 4 │ 2 │ 1 │
│...│...│...│
└───┴───┴───┘
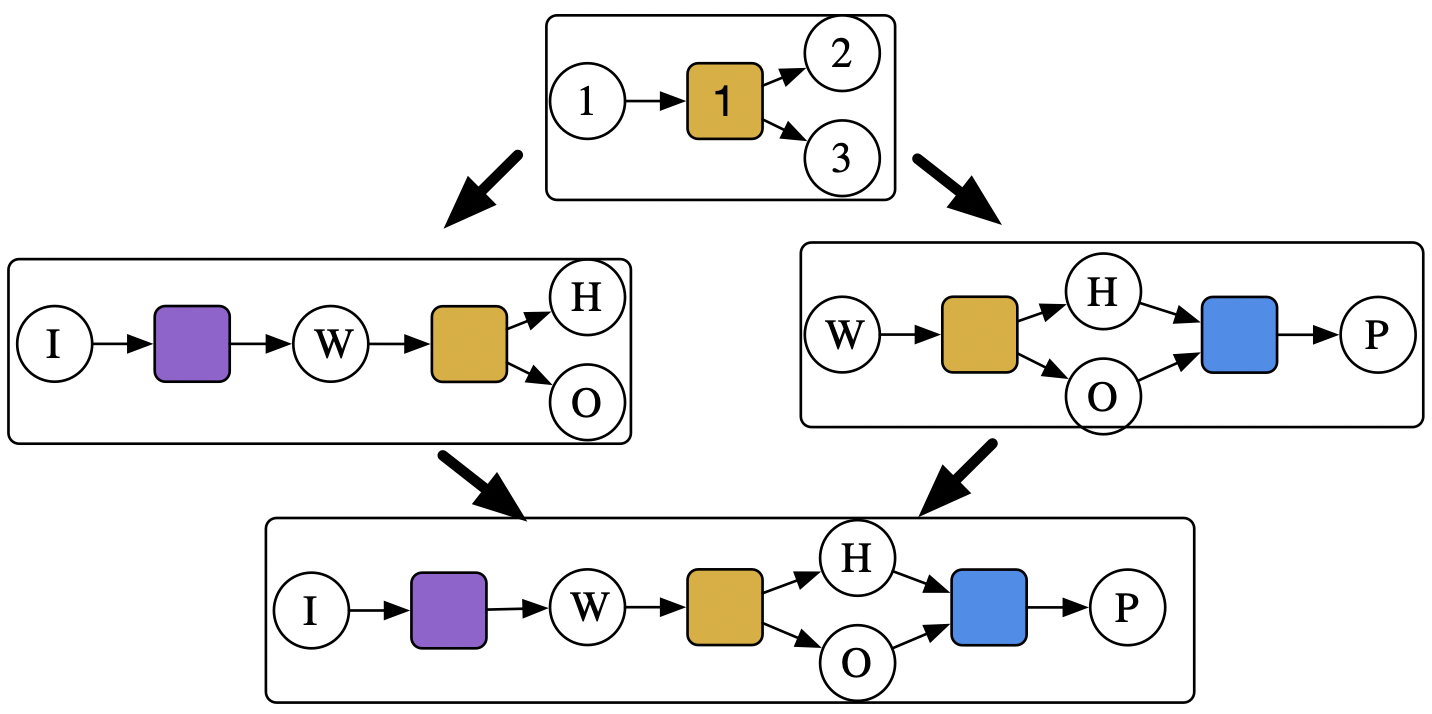
4 rows omitted💭 Colimits of CSets: gluing models together
Write a program that combines overlapping reaction networks:
- e.g. \(\boxed{\text{H}_2\text{O}_s\overset{melt}\rightarrow \text{H}_2\text{O}_l\overset{lyse}\rightarrow \text{H}_2+\text{O}_2}\) and \(\boxed{\text{Water}\overset{zap}\rightarrow \text{H gas}+\text{O gas}\overset{oxidize}\rightarrow Peroxide}\)
- This has one overlapping transition and three overlapping species
# Python
def merge(n1:RxnNet, n2:RxnNet,
s_overlap:list, t_overlap:list):
n1_rxn = deepcopy(n1.rxns)
for r2 in n2.rxns:
n_pairs = [(r1.name,r2.name) for r1 in n1_rxn]
if not intersect(t_overlap, n_pairs):
n1_rxn.append(rename_state(r2, s_overlap))
return RxnNet(n1_rxn)
def rename_state(r::Rxn, s_overlap):
s_dict = dict([(v,k) for (k,v) in s_overlap])
Rxn(r.name, rename_species(r.i, s_dict),
rename_species(r.o, s_dict))
def rename_species(s::Species, s_dict::dict):
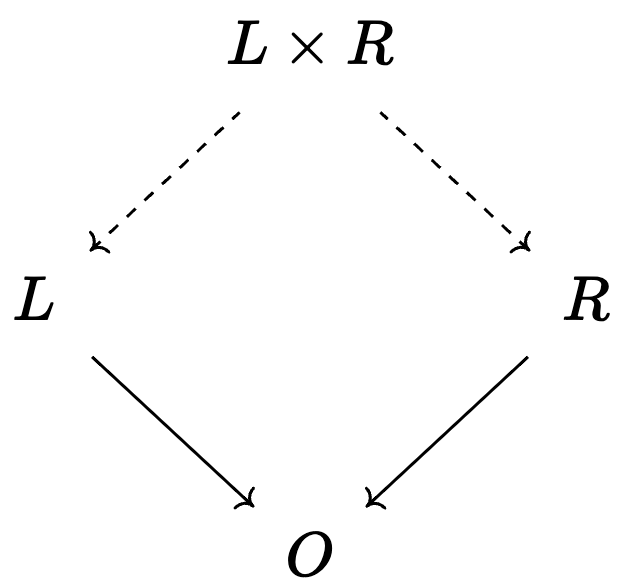
Species(get(s_dict,s.name, s.name))💭 Limits of CSets: model multiplication
Write a program that multiplies all stoichiometry by 2:
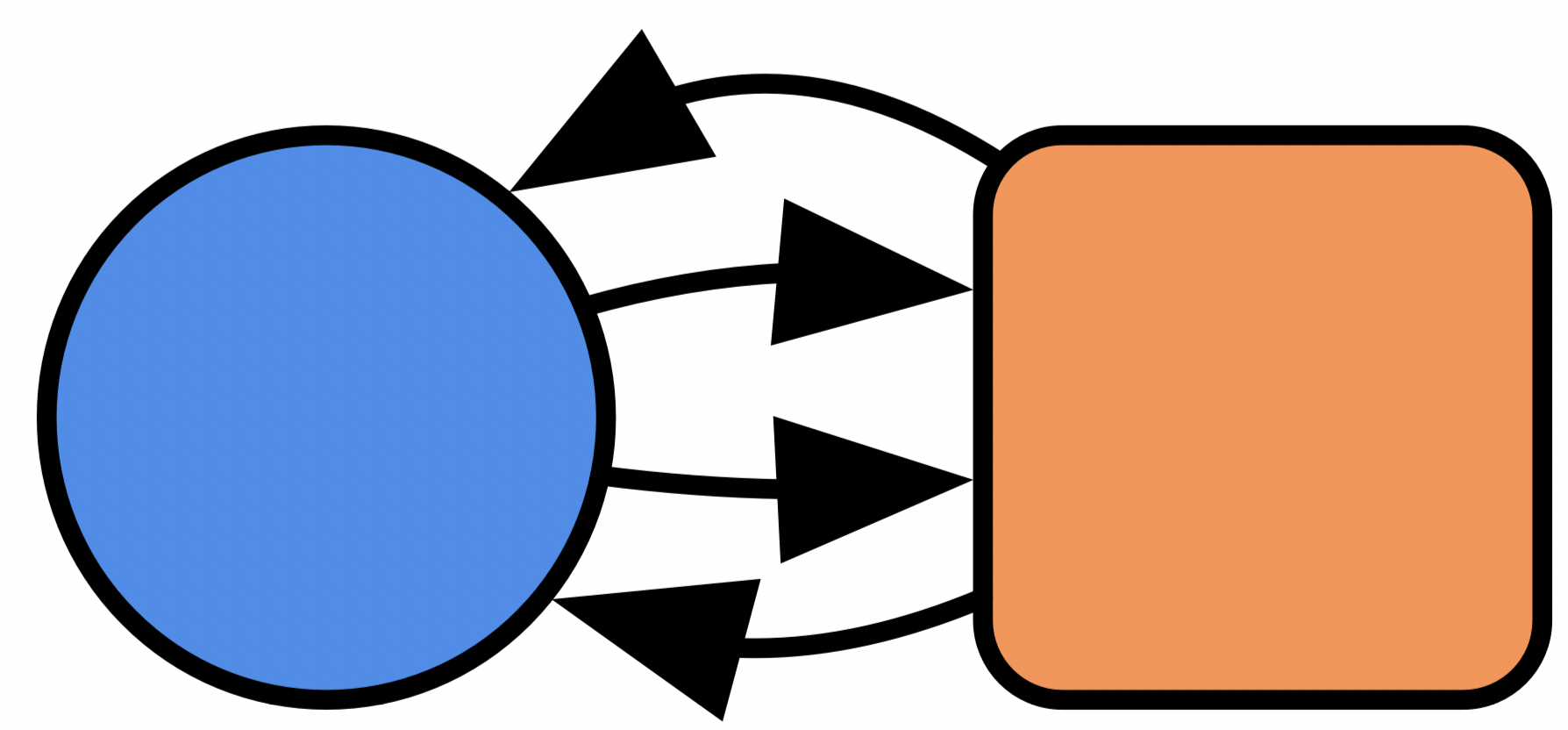
Write a program that stratifies a unary CRN with a CRN of phase transitions:
- e.g. stratifying \(A\rightarrow B\) with \(Solid \rightarrow Liquid \leftrightarrows Gas\) yields:
💭 Limits of CSets: model multiplication
Write a program that stratifies a unary CRN with a CRN of phase transitions:
- e.g. stratifying \(A\rightarrow B\) with \(Solid \rightarrow Liquid \leftrightarrows Gas\)
# Python
def strat(r1: RxnNet, r2:RxnNet):
rs = []
for rx1 in r1.rxns:
for s2 in get_states(r2):
rs.append(rename_rxn(rx1,s2))
for rx2 in r2.rxns:
for s1 in get_states(r1):
rs.append(rename_rxn(rx2,s1))
return RxnNet(rs)
def rename_rxn(r:Rxn, s: Species):
return Rxn(r.name, rSt(r.i,s), rSt(r.o,s))
def rSt(st: State, s2:Species)
return State(Dict([Species(s1.name+s2.name)
for (s1,v) in st.species.items()]))
def get_states(r::RxnNet):
...
💭 Takeaways
Gluing together models and multiplying models can be made sense of in any category.
- These operations often correspond to our genuine conceptual breakdown of complex systems
- This is not made explicit in conventional programming.
- These can be efficiently implemented for ACSets/databases generally in a standard library, so that custom code need not be written for each domain.
Outline
- 🗄️ Data
- CT: C-Sets, Homomorphisms, Data migration
- Examples: Computational chemistry simulation generated data
- 💭 Models
- CT: Colimits, Limits
- Examples: COVID models, Chemical Reaction Networks
- 📈 Simulation
- CT: Functors, DWDs, rewriting
- Examples: Multiphysics PDEs, Agent-based models
📈 Continuous time simulations
Simulations shouldn’t be written as code. The model should be compiled to simulation.
Code is a good semantics category, but lousy for syntax (can’t do fancy things like limits and colimits in it).
# Python
def F(rho, v, MeshCoordi,facenodes,centroid): # advection flux coeff
return -rho*(v @ fnormal(MeshCoordi,facenodes,centroid))
def get_linMatrix(pymesh, custom_mesh, dt, phi0):
. . .
totalMeshCells=len(MeshCells)
fluxMat=numpy.zeros((totalMeshCells, totalMeshCells))
bMat=numpy.zeros((totalMeshCells,1))
for c_cell in range(totalMeshCells):
n_index=0
for n_cell in neighbourID[c_cell]: #n_cell neighbouring cell index
c_ni_faceNodes=cellFaceID[commonFace[c_cell,n_index]]
if n_cell == None: #this will handle the boundary cell elements
fluxMat[c_cell,c_cell], bMat[c_cell]=get_Bcondition(...)
else: #non boundary elements
#D is diffusion flux contribution
D=gamma*gDiff(MeshCoordi,c_ni_faceNodes,[...])
#A is advection flux contribution
A=F(rho,numpy.array([ux,uy]),MeshCoordi,c_ni_faceNodes,[...])
fluxMat[c_cell,n_cell]=-(D*funA(A,D) + max(A,0)) #General scheme by Patankar
n_index+=1
bMat[c_cell]+=rho*cellvolume[c_cell]*phi0[c_cell]/dt
fluxMat[c_cell,c_cell]+= -( numpy.sum(fluxMat[c_cell,:]) - fluxMat[c_cell,c_cell] ) + rho*cellvolume[c_cell]/dt
return fluxMat, bMatConceptual model:
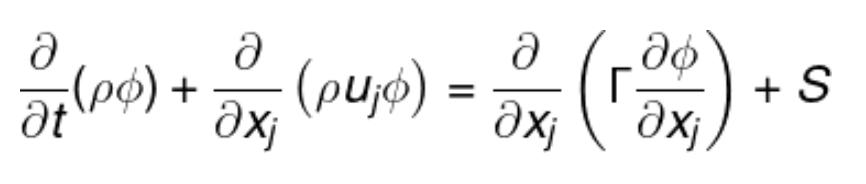
Problem
Implementation details to worry about when we update the math
- how much does order matter?
- which
forloops need modification? - which parts are geometry vs physics?
📈 Multiphysics modeling
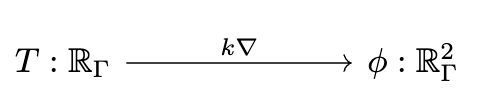
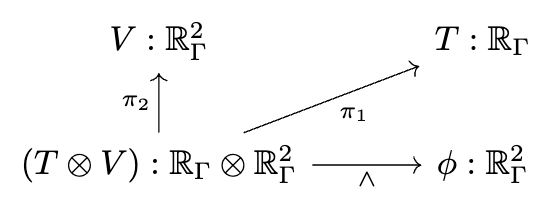
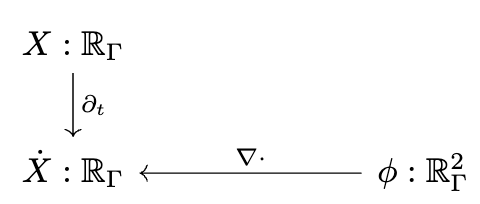
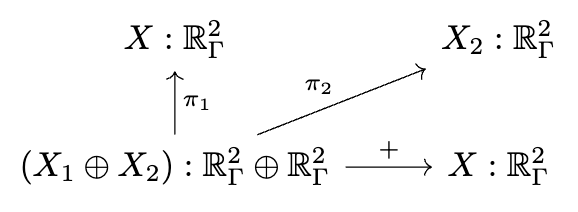
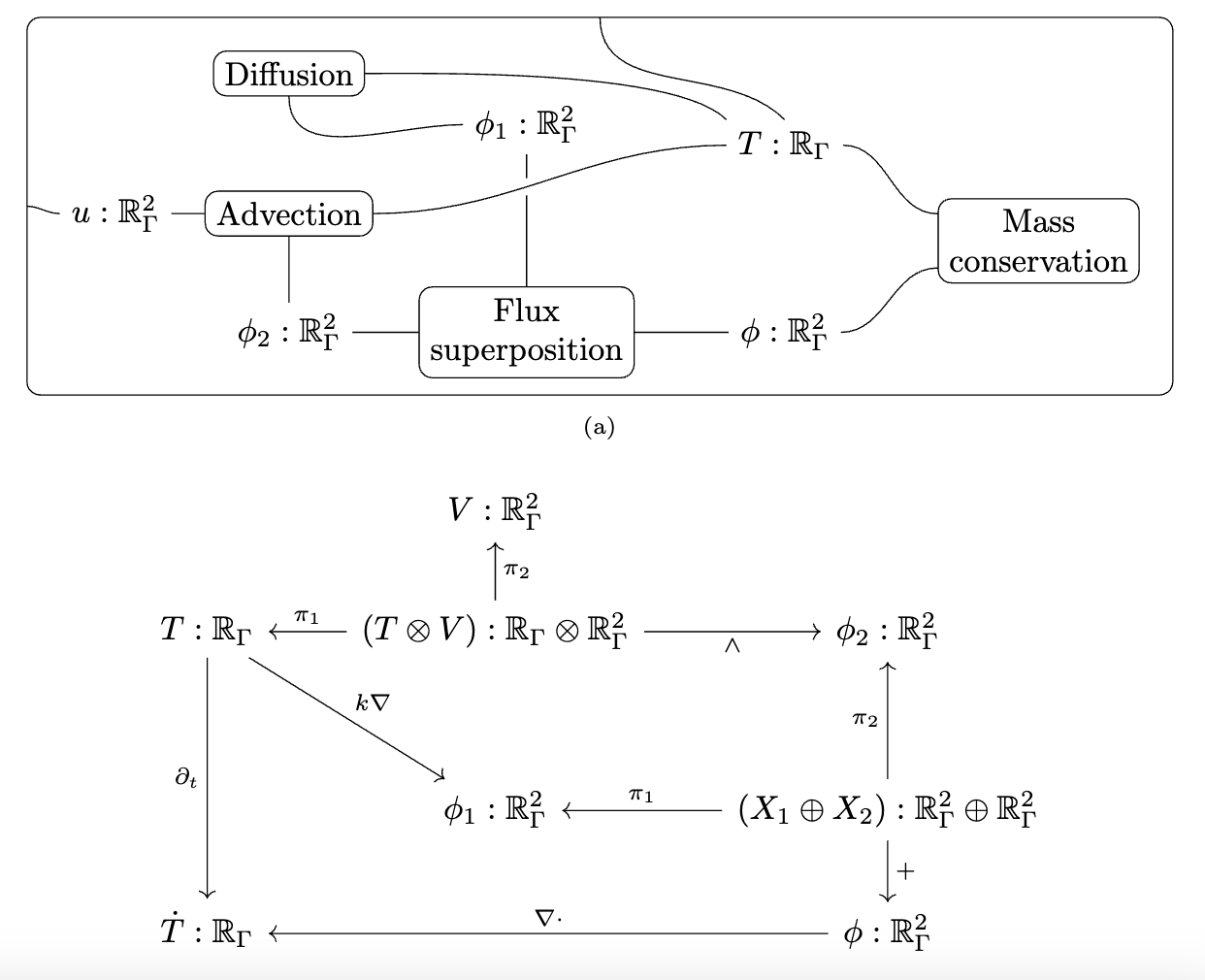
# AlgebraicJulia
"""Define the multiphysics"""
Diffusion = @decapode DiffusionQuantities begin
C::Form0{X}
ϕ::Form1{X}
ϕ == k(d₀{X}(C)) # Fick's first law
end
Advection = @decapode DiffusionQuantities begin
C::Form0{X}
(V, ϕ)::Form1{X}
ϕ == ∧₀₁{X}(C,V)
end
Superposition = @decapode DiffusionQuantities begin
(C, Ċ)::Form0{X}
(ϕ, ϕ₁, ϕ₂)::Form1{X}
ϕ == ϕ₁ + ϕ₂
Ċ == ⋆₀⁻¹{X}(dual_d₁{X}(⋆₁{X}(ϕ)))
∂ₜ{Form0{X}}(C) == Ċ
end
compose_diff_adv = @relation (C, V) begin
diffusion(C, ϕ₁)
advection(C, ϕ₂, V)
superposition(ϕ₁, ϕ₂, ϕ, C)
end"""Assign semantics to operators"""
funcs = sym2func(mesh)
funcs[:k] = Dict(:operator => 0.05 * I(ne(mesh)),
:type => MatrixFunc())
funcs[:⋆₁] = Dict(:operator => ⋆(Val{1}, mesh,
hodge=DiagonalHodge()), :type => MatrixFunc());
funcs[:∧₀₁] = Dict(:operator => (r, c,v)->r .=
∧(Tuple{0,1}, mesh, c, v), :type => InPlaceFunc())📈 Continuous time simulation
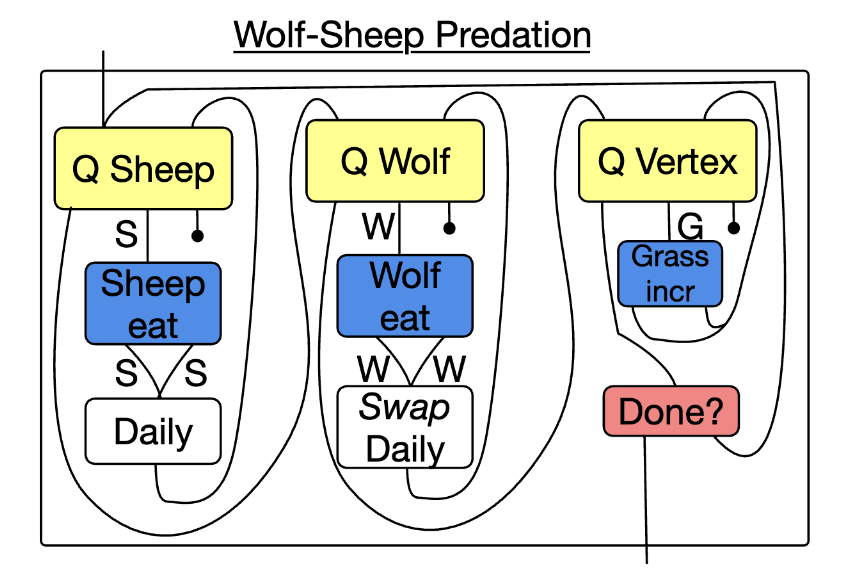
📈 Agent based models
# Python
class Wolf():
def init(self, eng: int, position):
def move(self):
...
class Sheep():
def init(self, eng: int, position):
def move(self):
...
class Grass():
def init(self, eng: int, position):
def increment(self):
...
class Graph():
def init(self, vertices, edges):
class World():
def init(self, graph, ws: list[Wolf], ss: list[Sheep]):📈 Merging, deleting, copying, adding … without code
Task: find a reaction \(A + B \rightarrow C\), delete \(C\) from the reaction network (if no other reactions need it), merge \(A\) and \(B\) together into \(AB\), and add a reaction \(\varnothing \rightarrow AB\)
# Python
def merge_delete_add(r:RxnNet):
# try to rewrite each reaction
for (i,rxn) in enumerate(r.rxns):
# determine if has at least two inputs
if sum(rxn.i.species.values()) < 2:
continue # cannot rewrite this rxn
# determine if we can delete an output
C = None
for out_species in rxn.o.species.keys():
appears_in = [j for (j, jrxn) in enumerate(r.rxns)
if out_species not in (jrxn.i.species
| jrxn.o.species)]
if appears_in == [i]:
C = out_species
if C is not None: # we can delete an output
out_state = State(Dict([(k,v-1 if k == C else v)
for k,v in rxn.o.species.items()]))
# . . . etc.
# to do: merge two input states
# to do: add a rxn that creates that merged state- What if pattern was changed?
- What if more than one reaction?
- What if reactions merged?
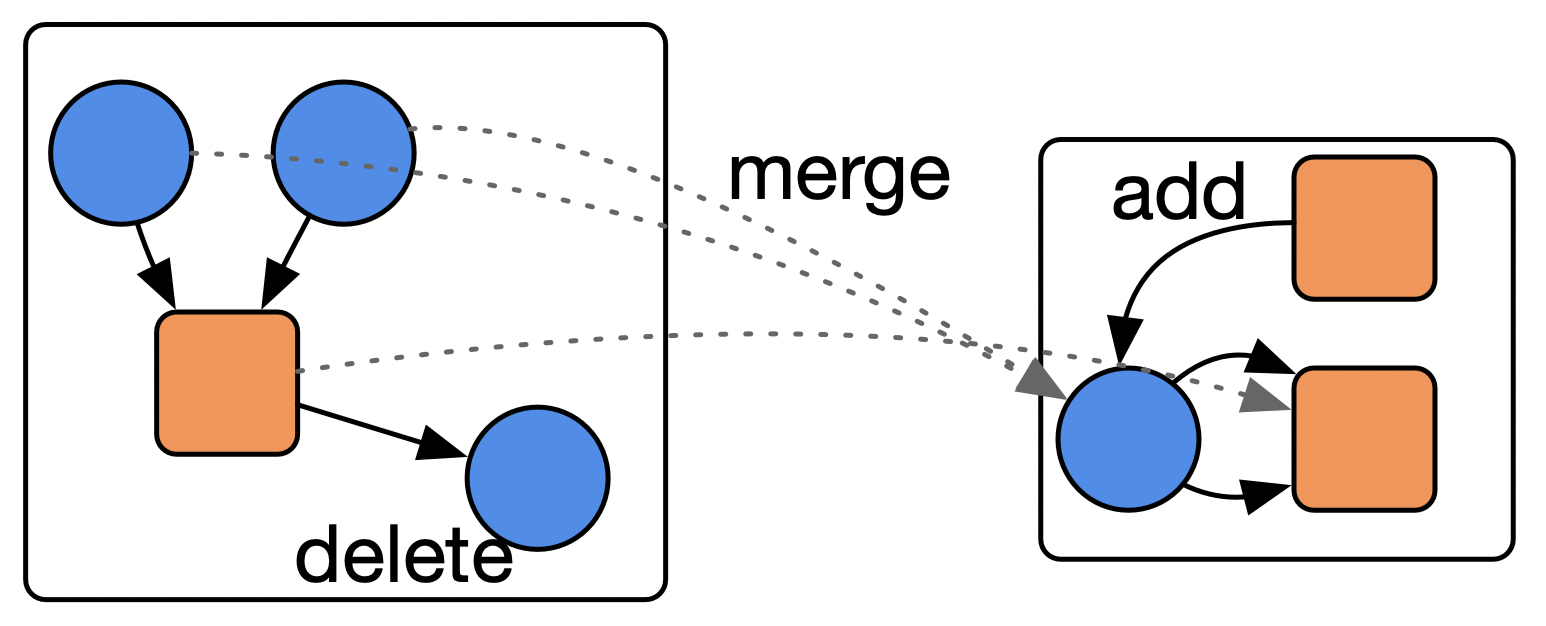
# AlgebraicJulia
Pattern = @acset PetriNet begin
S=3; T=1; I=2; O=1; it=1; ot=1; is=[1,2]; os=[3]
end
# Subobject of Pattern which we do not delete
I = @acset PetriNet begin S=2;T=1;I=2;it=1;is=[1,2];end
Replacement = @acset PetriNet begin
S=1; T=2; I=2; O=1; it=1; ot=2; is=1; os=1
end
rule = Rule(homomorphism(I,Pattern),
homomorphism(I,Replacement))
rewrite(rule, my_rxnet) # uses colimits underneath!Conclusions


Lack of automation due to unclear context and assumptions: models are not explicit.
Formalization creates possibility of automation - important as science scales both in amount of data and conceptual complexity of the data.
Conclusions
CT is useful for the same reason interfaces are generally useful. In particular, CT provides generalized notions of
- multiplication / multidimensionality
- adding things side-by-side
- gluing things along a common boundary
- looking for a pattern
- find-and-replace a pattern
- parallel vs sequential processes
- Mad Libs style filling in of wildcards
- Zero and One
- A point
- “Open” systems
- Subsystems
- Enforcing equations
- Symmetry
These abstractions all fit very nicely with each other:
- conceptually built out of basic ideas of limits, colimits, and morphisms.
We can use them to replace a large amount of our code with high level, conceptual data.
Resources
- Papers and talks: algebraicjulia.org
- Blog posts: algebraicjulia.org/blog and topos.site/blog
- Code: github.com/AlgebraicJulia
- For software engineers: Blog post
- This talk: krisb.org/research#talks
Backup slides
🗄️. Why not structured?
Why don’t researchers often don’t make structure of data explicit?
Difficult to reap the benefits of structure
The data when sitting still may be structured, but the moment you have to do something with it, you have to load from the database back into ad hoc data structures and code.
- e.g. I want to do a kinetics simulation based on the data within the database
Difficult to maintain the structure
Structure codifies certain assumptions made - when those assumptions change, it can be a lot of work to make things right again.
- e.g. we assumed we were only working with orthorhombic crystal structures, things that touch the
Cellclass break once we go hexagonal.
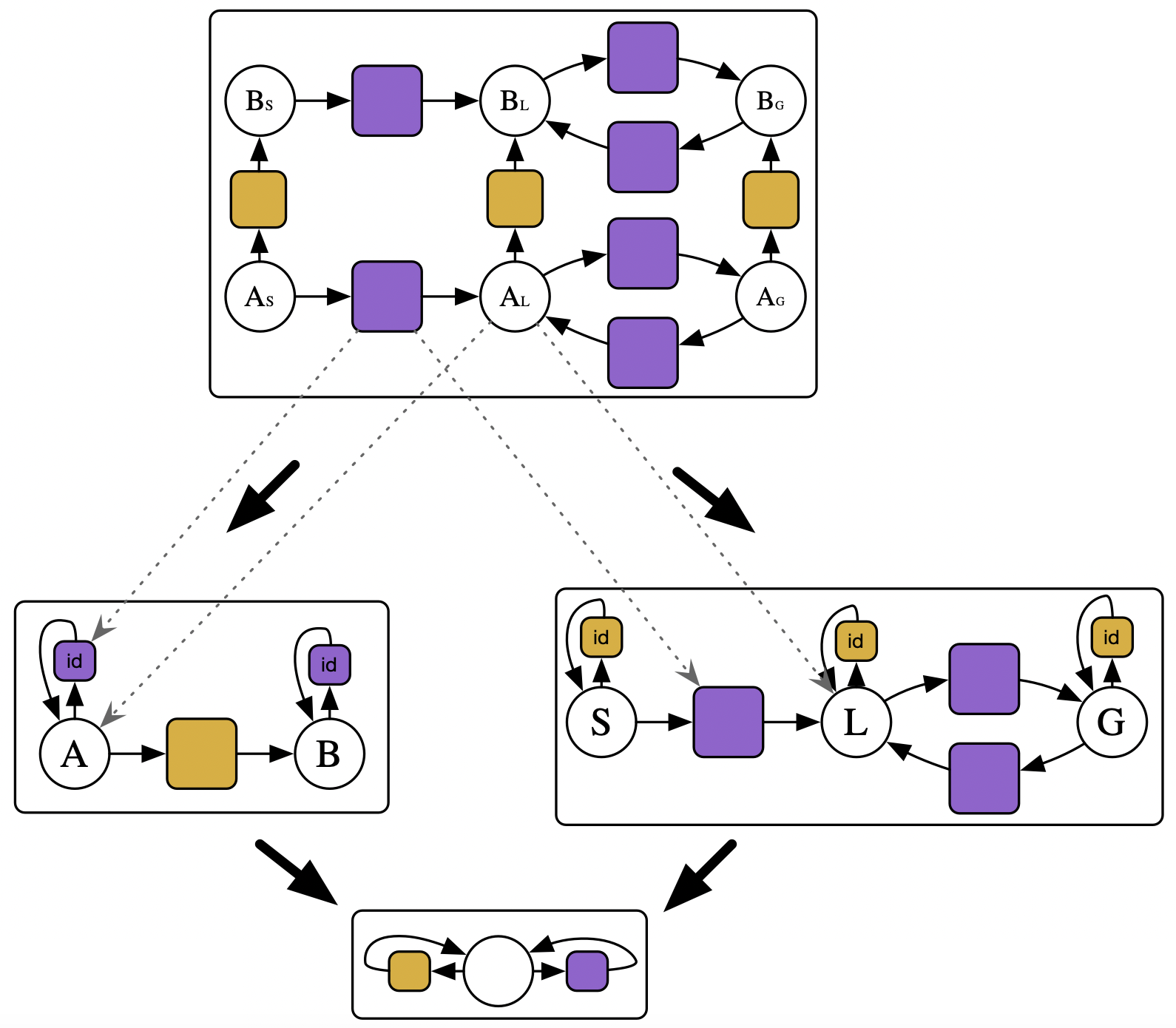
💭 Generic model stratification
"""Each S has a distinguished reflexive I, O, T"""
@present SchReflPetriNet <: PetriNet begin
refl_i::Hom(S,I)
refl_o::Hom(S,O)
refl_i ⋅ is == id
refl_o ⋅ os == id
refl_i ⋅ it == refl_o ⋅ ot
end
F = FinFunctor(SchPetriNet, SchReflPetriNet)base = @acset PetriNet begin
S=1; T=2; I=2; O=2;
is=1; os=1; it=[1,2]; ot=[1,2]
end
"""
Apply Σ;Δ migration to input X, then map into base
X transitions are sent to 1, added refl transitions to 2
(vice-versa if t = false)
"""
function strat_arr(X::PetriNet, t::Bool)
to_refl = ΣΔ(F)(X) # yields a morphism X → X′
Refl = codom(X_X′) # X, with reflective transitions
L = collect(to_refl[:T]) # set of original T's in X
# Determine where to send transitions in base
t_init = [p ∈ L ? t : !t for p in parts(Refl,:T)] .+ 1
homomorphism(Refl, base; initial=(T=t_init,))
end
function stratify(X::PetriNet, Y::PetriNet)
csp = Cospan(strat_arr(X, true), strat_arr(Y, false))
return Limit(csp)
end 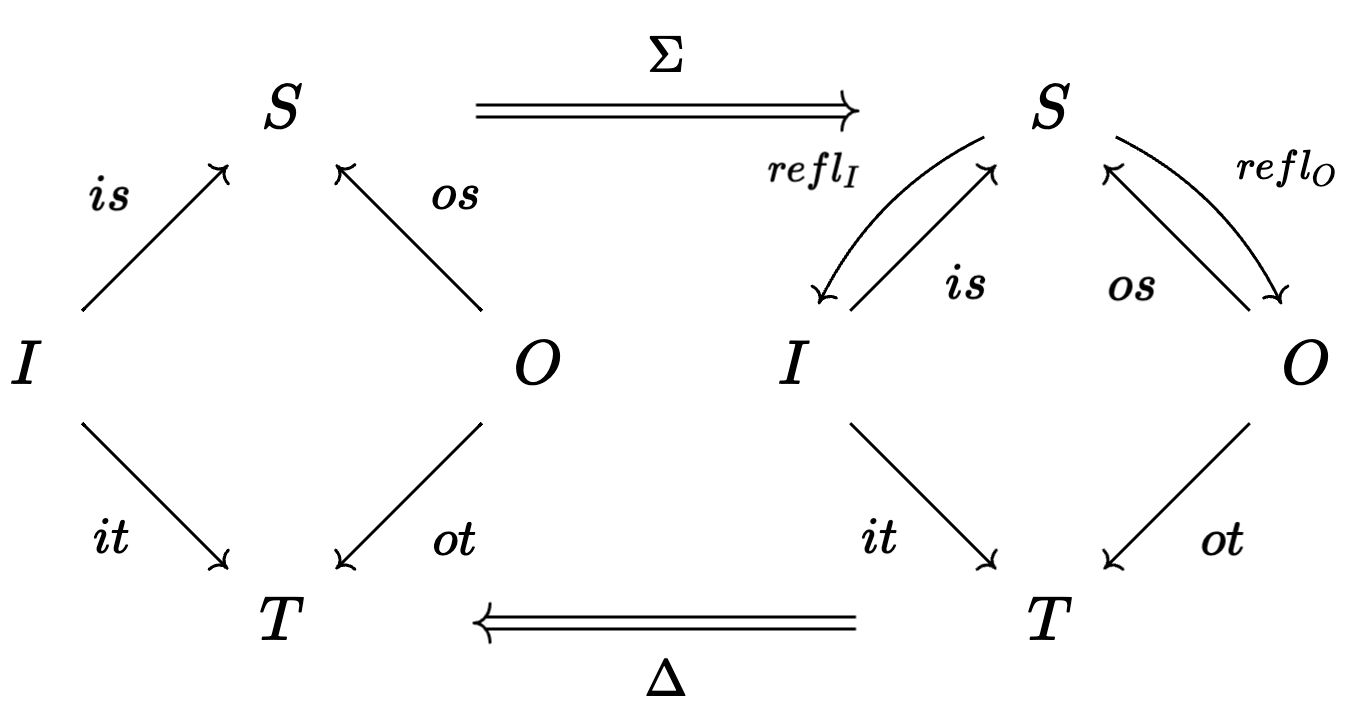

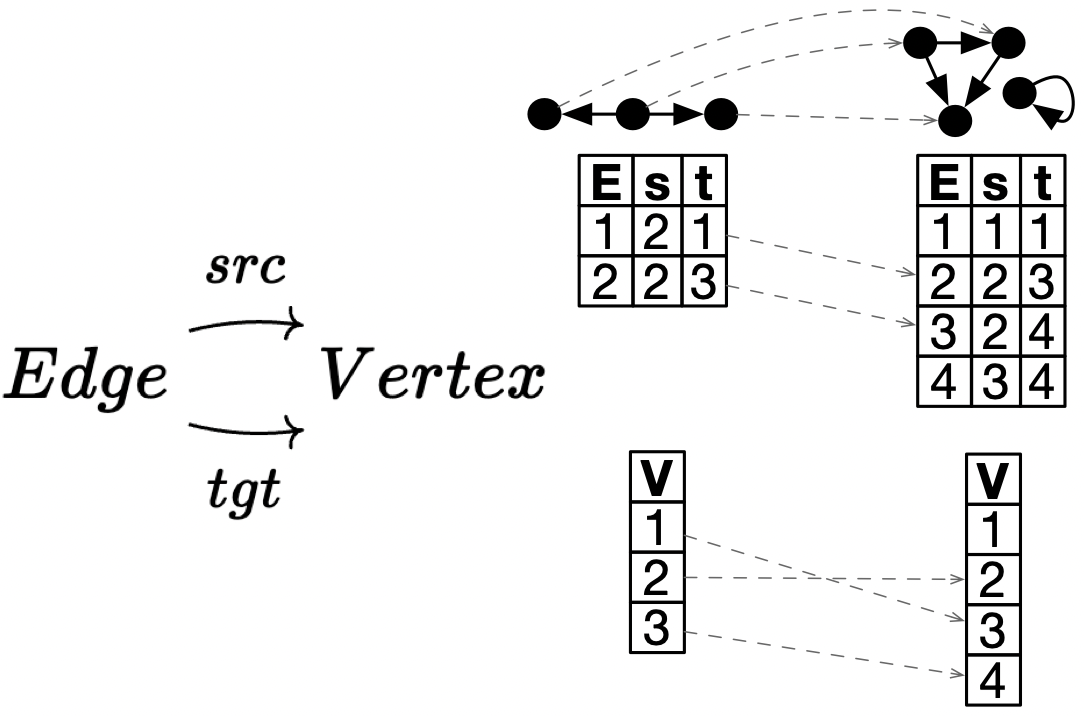
Graph morphism
About Me
Natural science
- 2015, Bachelors at Dartmouth: Physical chemistry / chemical engineering
- 2016, Visiting scholar at EPFL: organometallic synthesis
- 2016-2021, PhD at Stanford (Nørskov group)
- Applying Density Functional Theory to CO\(_2\) reduction catalysis
- Developing new DFT functionals for surface modeling
Computer science
- 2019-2020, Google: Software engineering intern
- 2020: Rotation in formal verification (Barrett Group)
Math + CS + Natural science
- 2022, UF: Postdoc in applied category theory (Fairbanks Group)
- 2023, Topos Institute: researcher, AlgebraicJulia developer
Misc content
“ACT helps you be more disciplined than you would be with just your intuitions.”
What’s wrong with the semantic web? It seeks a single language to represent everything. C.T. naturally resists this. SW is good at subtype hierarchies, but say you want to model a process - good luck! you might as well be encoding in JSON / XML … the logical framework isn’t helping.
Not a new standard (XKCD) but rather understanding
Why is C.T. pushout special? It helps us generalized beyond graphs
We want to say what the model is. Code is not model. (e.g. Imperial). The model in the scientist’s head may be complex (made up of many components) but it is not complicated (difficult for a human to grasp).
We have a toolbox of abstractions we understand rather than a universal language that can’t be extended once your assumptions change.
Our modeling framework latches onto the domain but we don’t have to start from nothing.